Post processing and visualization with PyVisA¶
In this example we are going to perform post processing on the methane hydrate system from one of the previous examples called “Using GROMACS”. All the post processing will be done through the features of PyVisA, that are found within the analysis tab of PyVisA, shown in Fig. 51, and the recalculation tool. We will continue the study of cage-to-cage diffusion within a S1 hydrate and add new collective variables. Then we will perform some cluster analysis, principal component analysis and visualization of the new results (NB: these methods of analysis requires the installation of the packages sklean and scipy).
Fig. 51 Analysis-tab of PyVisA with option for interactivity, animation and post processing.¶
Before we proceed with the analysis, we need to have some simulation results, so please run the example Using GROMACS first.
The example is structured as follows:
Visualization and compression¶
When the simulation has finished running, the results can be visualized and we can perform post processing of the data. It is always important to save the original data. This is because we are creating new data for PyVisA by using the trajectory files from the old simulation. By doing so there will be a loss of data equal to the frequency for which the trajectory files are stored. The following steps are suggested:
Compress the original data into a hdf5 file using:
pyvisa -i out.rst -cmp
Compress only the order parameter data into a hdf5.zip file using:
pyvisa -i out.rst -cmp -oo
Rename these files to standard_simulation.hdf5.zip
The results can be visualized by the rst-files, or by the compressed files. If you want to visualize the data, the following commands can be run:
pyvisa -i <input file>
pyvisa -i <rst-file> -data <data>
where <input file> can be either the rst-files or a hdf5.zip file. If you are using the -data command, then <input> can be ‘all’ which loads all the data, a trajectory file or a directory in the simulation. For example, the first path ensemble can be loaded by using 000 as <input>.
Through PyVisA you can visualize the trajectories from all path ensembles, and sort them based on their status and MC-move. Try finding and visualizing a reactive pathway by clicking on the plot, and in the Analysis-tab, press “Show trj”. This will highlight the points that belong to the selected trajectory. You can also customize how the visualization looks. In Fig. 52 a reactive pathway has been shown.
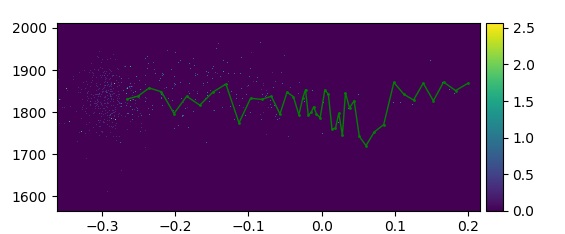
Fig. 52 Density plot of the orderparameter and the kinetic energy from the methane hydrate example with a reactive pathway shown in green.¶
Recalculation of new collective variables¶
Now that the old data has been stored we can add two more collective variables to the simulation using PyVisA. The collective variables we introduce are the area of the six-membered ring that the methane jumps through, and the volume of the starting cage. These descriptors will try to capture the breathing of the starting cage. The following lines must be added to the retis.rst file:
Collective-variable
-------------------
class = AreaAndVolume
module = orderp.py
With the collective variable added to retis.rst, we also need to add the Python script for the calculation in the orderp.py file. The Python package scipy is needed to run this script, so make sure that is is installed. The full script is given here:
Show/hide the Python script for the new collective variable. »
As we also have the z-coordinate of the methane molecule as a collective variable, we also need to add this to our orderp.py file so that the recalculation tool can use it. See the full script for the Position descriptor here:
Show/hide the Python script for the Position collective variable. »
Now, to begin the recalculation, we start PyVisA by loading all the data with the retis.rst file with the command:
pyvisa -i retis.rst
It is important to load PyVisA with the modified retis.rst file, as we need the program to know what has been added. Now we can access the feature for recalculation through the file-menu in PyVisA. Go to the file-menu, and press the option named “Recalculate data”. The following warning will show, Fig. 53, and press yes, if you have added the requirements for the post processing to both the retis.rst file and the orderp.py file.
Fig. 53 The warning issued by PyVisA before the recalculations.¶
The recalculation will now start. When the procedure is finished PyVisA will load the new data into the gui, and display a message letting you know that the new data can be visualized.
Post processing and clustering¶
Now that we have the new data, we can use the features of PyVisA to perform clustering, PCA. The following steps can be done in the data exploration:
- Go to the Analysis-tab of PyVisA and press the button “Show correlations”. This will produce the correlations matrix. From these results, plot the order parameter and one of the collective variables.
- Select a number of components to use in clustering and from the Analysis-selection, pick an algorithm for clustering and press the Analysis-button. This will produce a cluster plot of the chosen variables. Try to start with k-means, and then try Gaussian mixture and Spectral clustering to see if there is a difference between the methods.
- Try to perform a principal component analysis of the results. Begin by selecting 3 components, and PCA as the method, and press the analysis- button. This will produce the loading matrix, the scores plot from the first two components, and the cumulative explained variance. How much variance was retained? Where three components enough? Are there any strong correlation between the principal components and the original descriptors?
If you want to study the results from the principal component analysis further, the data will be stored in a hdf5 file in your simulation directory containing all the simulation data, and the data from the analysis.