Main studies performed with PyRETIS¶
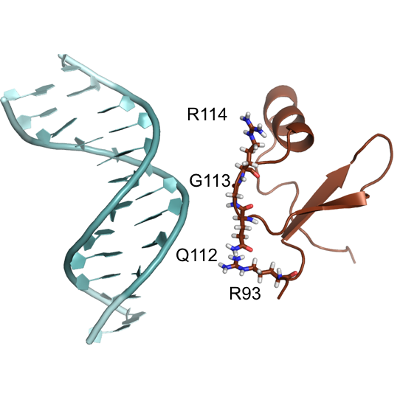
Predicting the mechanism and rate of H-NS binding to AT-rich DNA.
The adsorption of H-NS on DNA is studied at atomistic resolution with GROMACS. Local minima have been located by metadynamics and the transition rates computed by RETIS.
Paper: Predicting the mechanism and rate of H-NS binding to AT-rich DNA
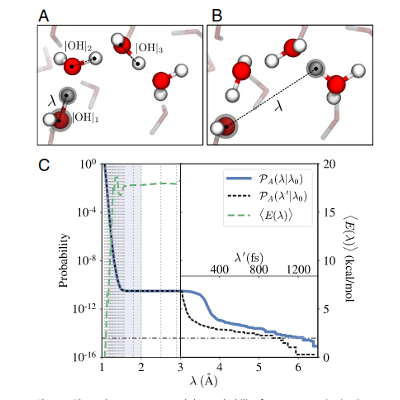
Autoionization of water.
BO-DFT simulations, via the RETIS approach, were used to study water autoionization. The mechanism(s) have been highlighted and their rate(s) quantified. Machine learning was applied to test the quality of the order parameters.
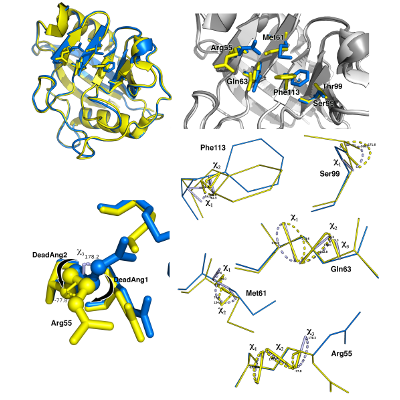
Conformational study of Cyclophilin - A.
Full atomistic simulations with GROMACS have been performed to sample and quantify the rate of the structural rearrangements of CyP-A and its muted confomer.
Paper in preparation.

Proton transfer in a water trimer.
A study on the proton transfer reaction with a polarizable potential is included. The various features of PyVisA can be tested on the simulation outputs.
Paper: PyVisA: Visualization and Analysis of path sampling trajectories
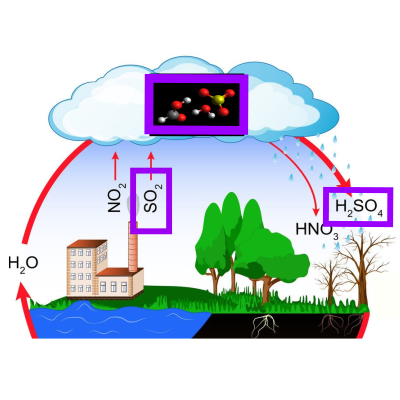
Formic acid catalysed formation of sulfuric acid.
A study on the formic acid catalysed conversion of sulfur trioxide and water to sulfuric acid. The mechanism(s) have been highlighted and their rate(s) estimated as a function of the temperature.
Paper: Path sampling for atmospheric reactions: formic acid catalysed conversion
Enhanced path sampling using subtrajectory Monte Carlo moves.
A study on the effect of using the subtrajectory Wire Fencing move compared to the standard shooting move. Systems studied are the 1D potential double-well, thin film breaking and a ruthenium redox reaction.
Paper: Enhanced path sampling using subtrajectory Monte Carlo moves
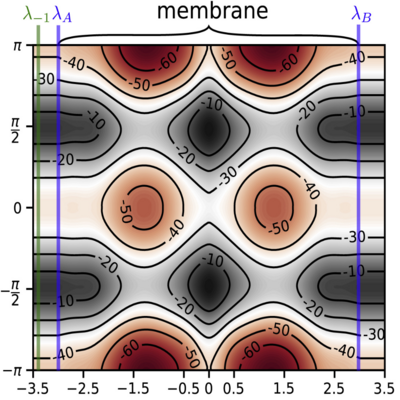
Memory reduction using partial path ensembles.
A study on how the path ensemble definitions can be changed when long-lived metastable states are present in the reaction.
Paper: Path sampling with memory reduction and replica exchange to reach long permeation timescales